Bio
I am a finishing PhD student at the University of Vermont, USA. I am interested in artificial life, the evolution of evolvability and development.
My PhD aims to investigate evolutionary dynamics in variable environments using computational models and whole-genome sequence analysis. My main results highlight the importance of the interaction between the genotype-phenotype map and the fitness landscape. In my models of gene regulatory networks, this interaction determined the effect environmental variability had on the evolving populations. Similarly, genetic variation involved in local adaptation to pH variability in sea urchin populations were found in genomic regions influencing the genotype-phenotype map.
Published research
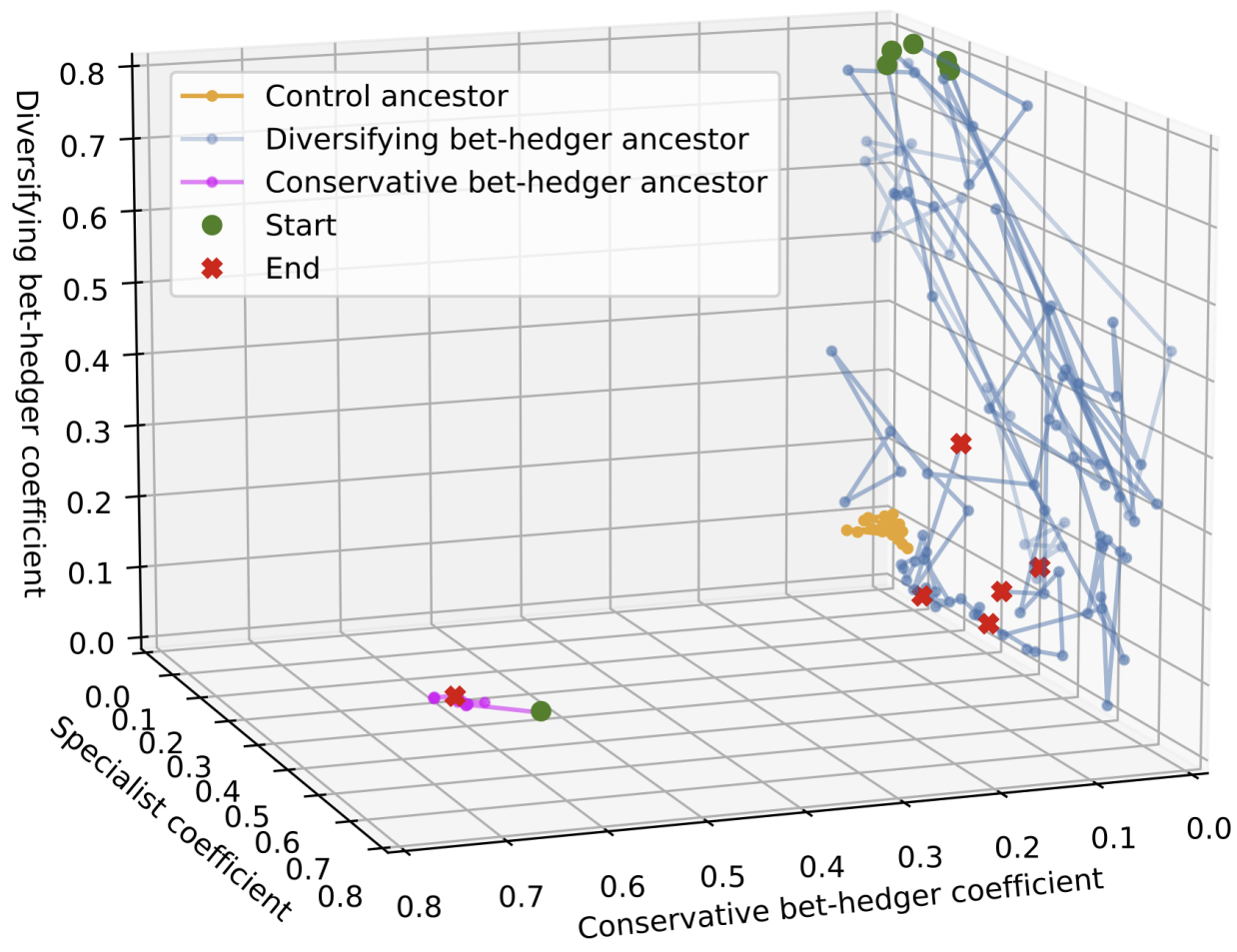
- Simple gene regulatory networks in a variable environment first evolved such that a few mutations could result in switching between phenotypes optimal in alternative environments - diversifying bet-hedgers reduced risk by making sure at least some of their offspring would be fit regardless of an unexpected environmental change. However, we found this strategy was quickly outcompeted by a more conservative strategy that only produced offspring with a phenotype intermediate between the two alternative optimal phenotypes. We think that this was due to the properties of the genotype landscape underlying these strategies. Published peer-reviewed paper.
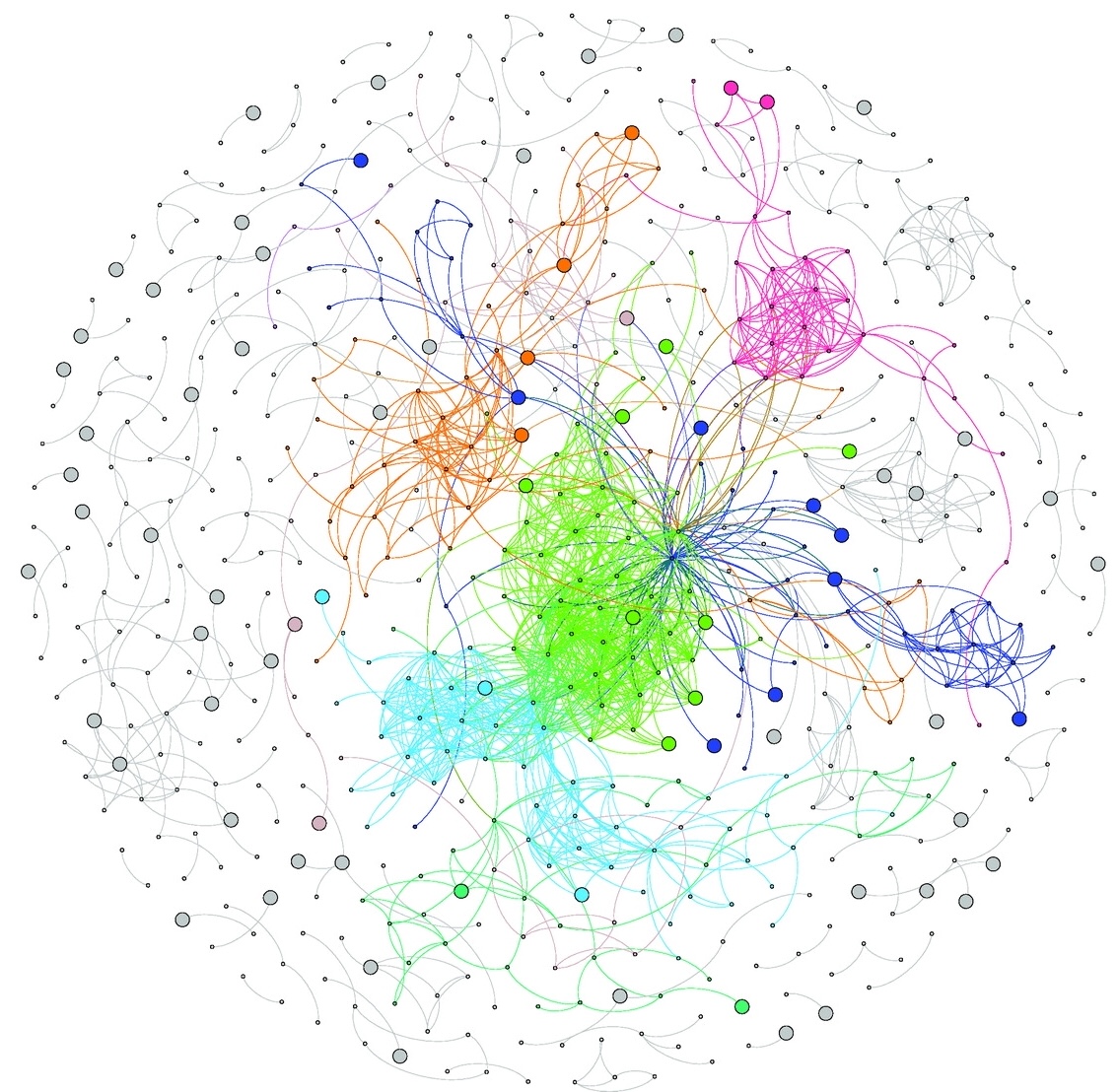
- By whole-genome sequencing a 140 purple sea urchins collected from seven populations experiencing different levels of environmental variability, I generated a very useful resource to investigate the structure and genetic diversity of these marine populations, as well as to find genetic variation putatively involved local adaptation to environmental variability. Interestingly, we found that such genetic variation was mostly regulatory and found in regions predicted to have phenotypic effects with low levels of pleiotropy. Published peer-reviewed paper.
Class final projects
Foundations of Quantitative Reasoning pdf
Title: The effects of linkage disequilibrium on maintaining balanced polymorphisms in seasonal adaptation
Evolutionary Computation pdf
Title: Comparing different implementations of the classical gene regulatory network evolutionary model: the start of an exploration
Principles of Complex Systems pdf
Title: The evolution of gene regulatory networks in variable environments
Data Science I pdf
Title: Signatures of adaptation to environmental variability in the protein-protein interaction network
Resources
Whole-genome sequence data available here: BioProject PRJNA674711
Protocol for microinjection of reporter constructs into sea urchin fertilized eggs
General urchin care information: Filter changes, Cleaning/feeding, Technical set-up
